
Audio File Formats Explained (Mp3 vs Wav vs AIFF vs FLAC & More)
The IGV session XML file is prepopulated with track files generated by DRAGEN.The session file loads the reference genome that best matches the standard reference genomes in an IGV installation, by comparing the name of the--ref-dir specified on the command-line. Standard UCSC human reference genomes are autodetected, but any variations from the standard reference genomes might not be.

How To Work with JSON in JavaScript Sasher Pakistan
To see an example of turning a bedDetail custom track into the bigBed format, see this How to make a bigBed file blog post. PSL format. PSL lines represent alignments, and are typically taken from files generated by BLAT or psLayout. See the BLAT documentation for more details. PSL data tracks can also be visualized in rearrangement display.
pyDNase Analysis Scripts — pyDNase 0.2.4 documentation
Create a wig format file following the directions here. Note that when converting a wig file to a bigWig file, you are limited to one track of data in your input file; you must create a separate wig file for each data track. Note that this is the file that is referred to as input.wig in step 5 below.

Hair wig PNG transparent image download, size 3094x2392px
The wiggle (WIG) format is for display of dense, continuous data such as GC percent, probability scores, and transcriptome data.. Wiggle format is composed of declaration lines and data lines.. These formats were developed to allow the file to be written as compactly as possible.

Advantages and Disadvantages of Popular Audio Formats Part 2
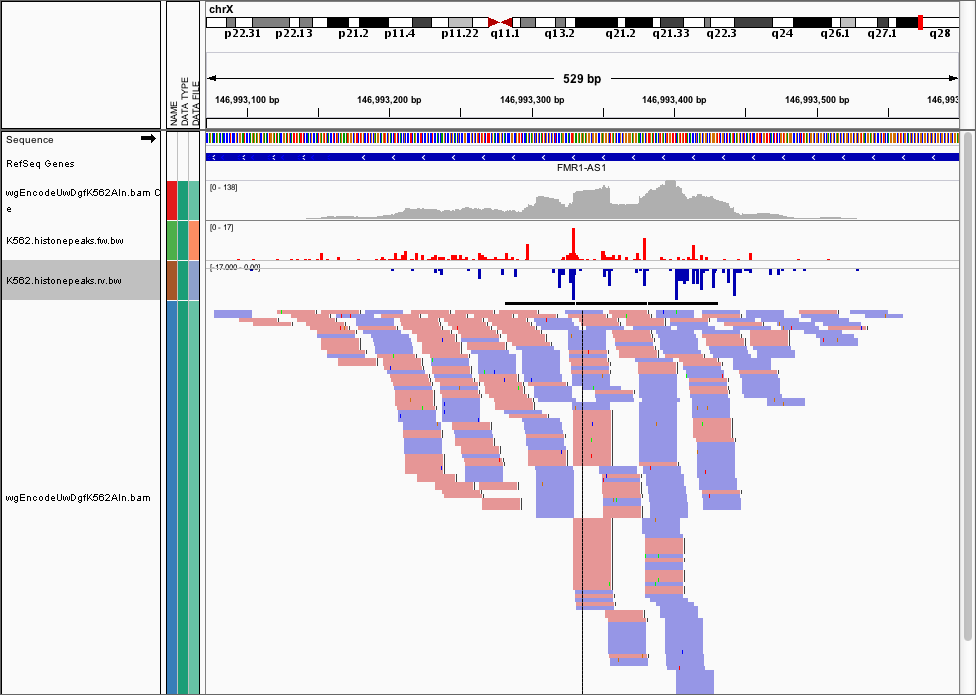
Abstract. BigWig files are a compressed, indexed, binary format for genome-wide signal data for calculations (e.g. GC percent) or experiments (e.g. ChIP-seq/RNA-seq read depth). bwtool is a tool designed to read bigWig files rapidly and efficiently, providing functionality for extracting data and summarizing it in several ways, globally or at specific regions.
/GettyImages-181944682-58e3af0a3df78c5162007a81-3cf8499c59d7463fac41e50bb9409103.jpg)
Best Graphics File Formats for Desktop Publishing
The bigWig format is for display of dense, continuous data that will be displayed in the Genome Browser as a graph. BigWig files are created initially from wiggle (wig) type files, using the program wigToBigWig. The resulting bigWig files are in an indexed binary format.
How to Show File Extensions on iPad & iPhone in Files App
A WIG file (.wig) is a text file that defines either a feature or data track. The WIG file format is described on the UCSC website. Required extension .wig. For faster loading, convert WIG files to bigWig format. Alternatively, convert to TDF format using igvtools.

Hair wig PNG
BioJS is an open source JavaScript library of components for the visualisation of biological data on the web. Here we present wigExplorer, a standard, portable BioJS component designed to easily render wig data format files. wigExplorer can be integrated and controlled from other applications. To our knowledge, this is the first modular.

Hair wig PNG
What is bigWig format? BigWig is a file format for display of dense, continuous data in a genome browser track, created by conversion from Wiggle ( WIG ) format. BigWig format is described at the UCSC Genome Bioinformatics web site , and the Broad Institute file format guide provides additional information.
How to file extension caqwecosmetics
Wiggle Track ASCII Text Format (.wig) Wiggle files and its bedgraph variant allow you to plot quantitative data as either shades of color (dense mode) or bars of varying height (full and pack mode) on the genome.. The bedGraph format, like all BED-based formats and most file formats used by UCSC, use "0-start, half-open" coordinates, but the.

Hair wig PNG transparent image download, size 1000x1244px
con: A path, URL, connection or WIGFile object. For the functions ending in .wig, the file format is indicated by the function name.For the base export and import functions, the format must be indicated another way. If con is a path, URL or connection, either the file extension or the format argument needs to be "wig". Compressed files ("gz", "bz2" and "xz") are handled.

Understanding video File , Standard , Extension , File , Format YouTube
To display data as bar graphs along the genome e.g. in the UCSC genome browser, you can create BigWig files. The underlying Wig format is described in more detail here. BigWig is the binary version (described here), that allows compressing the data and streaming of the data from a remote location to the machine running the display (i.e. the genome browser) on demand.

Common image file formats and when to use them Creative Bloq
These formats were developed to allow the file to be written as compactly as possible. The bigWig format is an indexed binary form of the wiggle file format, and is useful for large amounts of dense and continuous data to be displayed in a genome browser as a graphical track. As mentioned above, the visual representation of this format is very.

Wig PNG Transparent Images PNG All
BigWig files are a compressed, indexed, binary format for genome-wide signal data for calculations (e.g. GC percent) or experiments (e.g. ChIP-seq/RNA-seq read depth). bwtool is a tool designed to read bigWig files rapidly and efficiently, providing functionality for extracting data and summarizing it in several ways, globally or at specific regions.

Hair wig PNG transparent image download, size 1177x884px
Create a wig format file following the directions here. When converting a wig file to a bigWig file, you are limited to one track of data in your input file; therefore, you must create a separate wig file for each data track. Step 2. Remove any existing "track" or "browser" lines from your wig file so that it contains only data. Step 3.

Your Guide to File Types (and How to Convert Them) Onhax Me
This format is an alternative to wig and the bigwig formats and is typically used for data that has a value per chromosomal position, like for example coverage data. You can create TDF files directly from BAM files or from wig files. Creating TDF files (recommended) bam2tdf: convert read alignment to coverage plot